Research Projects
Mechanism of dopamine receptor activation
Dopamine receptors are important modulators of human physiology by responding to the endogenous neurotransmitter dopamine in the CNS. The family of dopamine receptors (D1 – D5) are also targets for several neurological disorders, including Parkinson’s and Alzheimer’s disease. The different dopamine receptor subtypes are diverse in structure, pharmacology, and transducer coupling (DRD1 and DRD5 couple primarily to Gs while D2-D4 signal by activating Gi). While the structures of DRD2, DRD3, and DRD5 bound to their antagonists have been determined, the structures of activated dopamine receptors were previously unresolved. In 2020, the Rosenbaum Lab reported the first active-state structure of the D2R-Gi complex reconstituted in a lipid bilayer by cryo-electron microscopy. As all GPCRs are embedded in the phospholipid bilayer membrane, it is important to understand how the lipid milieu modulates the receptor’s conformation and dynamics. Unlike previous detergent-bound complexes, our structure revealed a previously unappreciated role of lipids interacting with the complex, including the burial of the receptor’s helix 8 in the membrane. Future efforts will focus on using this structural knowledge to develop positive allosteric modulators for the D2 receptor.
Drug binding to the orexin receptors
The orexin receptors modulate circadian rhythms, feeding, and arousal, and are drug targets for sleep disorders (insomnia and narcolepsy), obesity, anxiety, and addiction. We determined the first molecular structures of both orexin receptor subtypes, OX1R and OX2R. Our results elucidated the mechanism of binding of the insomnia drug suvorexant, and explained the mechanism of subtype selective binding of the OX1R antagonist SB-674042. Our structural findings established how orexin receptor antagonists adopt a compact U-shape pose and bind deeply in the orthosteric pocket of the GPCR. Based on these findings, we have further enabled the structure-based design of selective orexin receptor antagonists. Our future aims are to determine the structure of the activated orexin receptors bound to agonist and G protein, which will illuminate the mechanism of neuropeptide activation and facilitate agonist drug design.
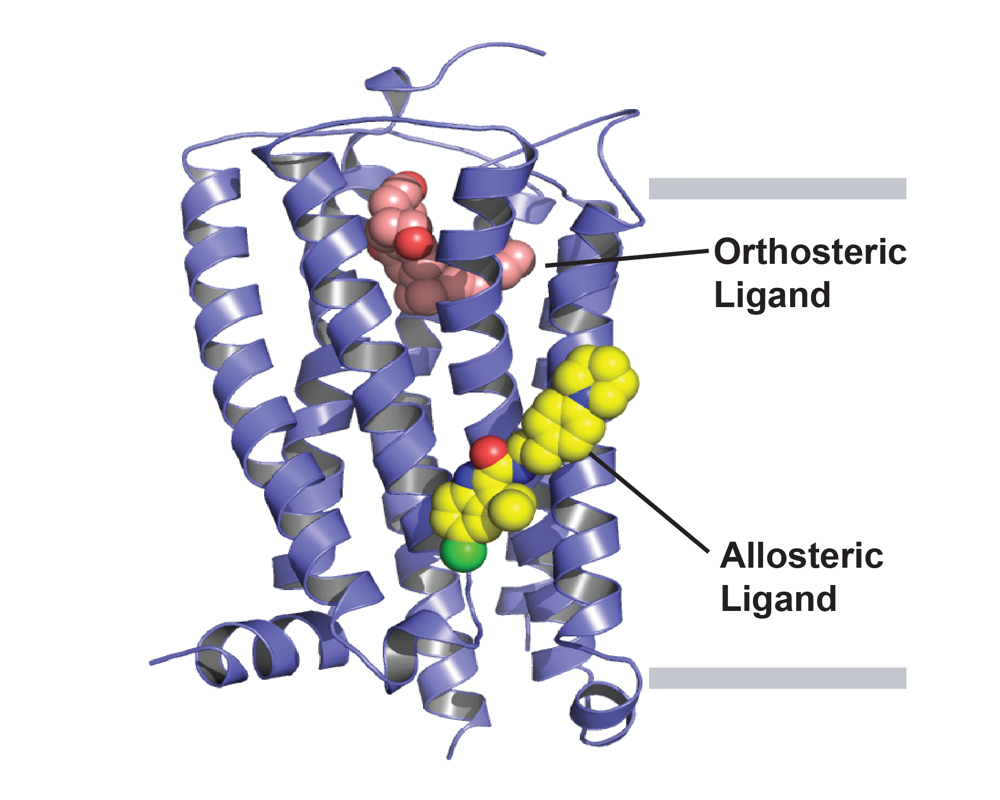
Allosteric modulation in the cannabinoid receptor CB1
CB1 is the most abundant GPCR present in the CNS and mediates functional responses to endocannabinoids and cannabis compounds. Beyond cannabis, CB1 is also a promising drug target for many CNS disorders, including epilepsy, pain, and anxiety. The Rosenbaum Lab determined the first high-resolution crystal structure of CB1 bound to the potent inverse agonist drug taranabant. Beyond such orthosteric inhibitors, allosteric modulators can finely tune the activity of GPCRs such as CB1, providing alternative therapeutic profiles. Our lab recently published the first structure of CB1 with an allosteric modulator, the compound ORG27569. This structure reveals an intermediate state of the receptor in an overall inactive confirmation despite the presence of an agonist in the orthosteric binding pocket. Our future aims are to understand how the compound cannabidiol works, both at CB1 and other receptors. Cannabidiol is a non-psychoactive compound present in Cannabis sativa, and was recently approved by the FDA for treating refractory epilepsy. Despite years of research, the molecular mechanism of CBD remains elusive, with weak affinity towards CB1 and the ability to modulate other targets. We are studying the mechanism of action of CBD using a combination of structural, pharmacological, and biochemical approaches.
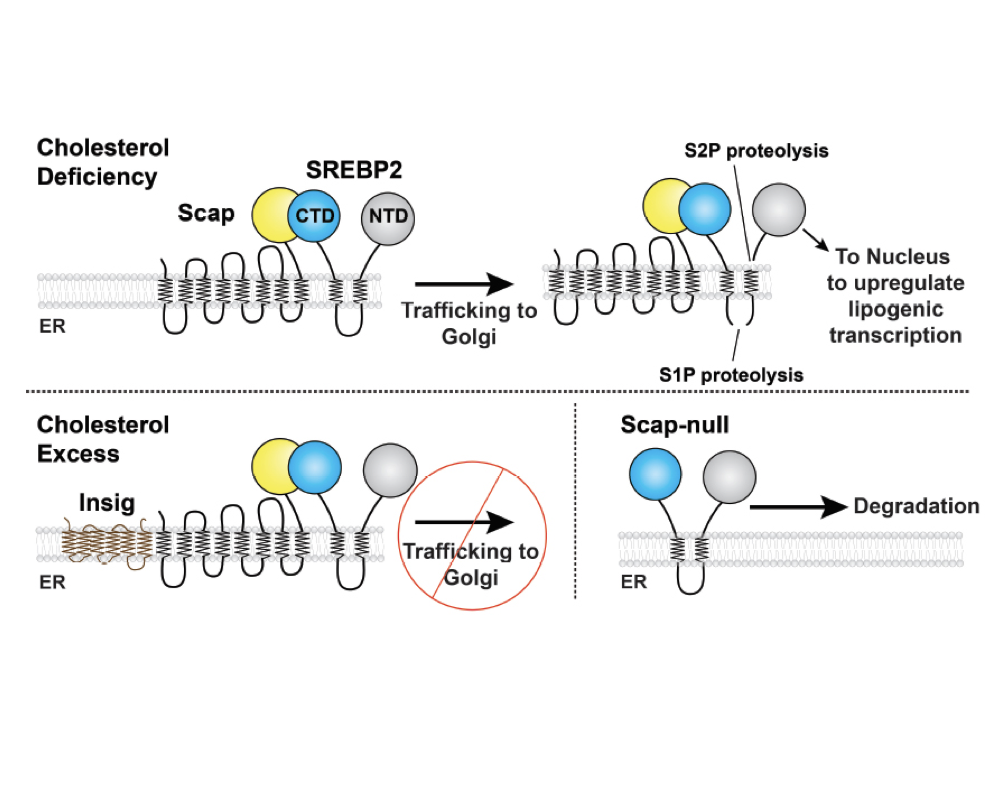
Molecular mechanism of sterol homeostasis
Cells must tightly regulate membrane lipid composition. Membrane cholesterol levels are sensed and regulated by Endoplasmic Reticulum (ER)-resident proteins of the SREBP pathway, including Scap, Insig, and SREBPs. Genes for cholesterol biosynthesis are controlled by SREBP2, whose processing is regulated by the concentration of cholesterol within the ER membrane. This ER membrane cholesterol content is directly sensed by the transmembrane protein Scap. When ER membrane cholesterol drops beyond a threshold of 5 mol percent, Scap transports the SREBP2 precursor from ER to Golgi, where SREBP2 is processed by membrane-embedded proteases. This process releases SREBP2’s transcription factor domain for nuclear import in order to upregulate cholesterol synthesis and uptake. Once cholesterol levels have risen above the threshold, Scap binds cholesterol and is also engaged by Insig-1 or Insig-2. These events cause Scap to adopt a non-trafficking conformation and retain SREBP2 in the ER. While the major steps of this pathway have been delineated, we lack a structural or biophysical understanding of the mechanisms of these proteins. We are pursuing atomic structures of the Scap-SREBP system proteins to understand 1) how Scap senses cholesterol and how cholesterol binding induces conformational changes to regulate its ER-Golgi localization; 2) how the binding of Scap to Insig enforces its non-trafficking conformation and ER retention; 3) how Scap interacts with SREBP2 in order to transport it to the Golgi. Finally, we are engaged in discovery of small molecule modulators of Scap-SREBP function.